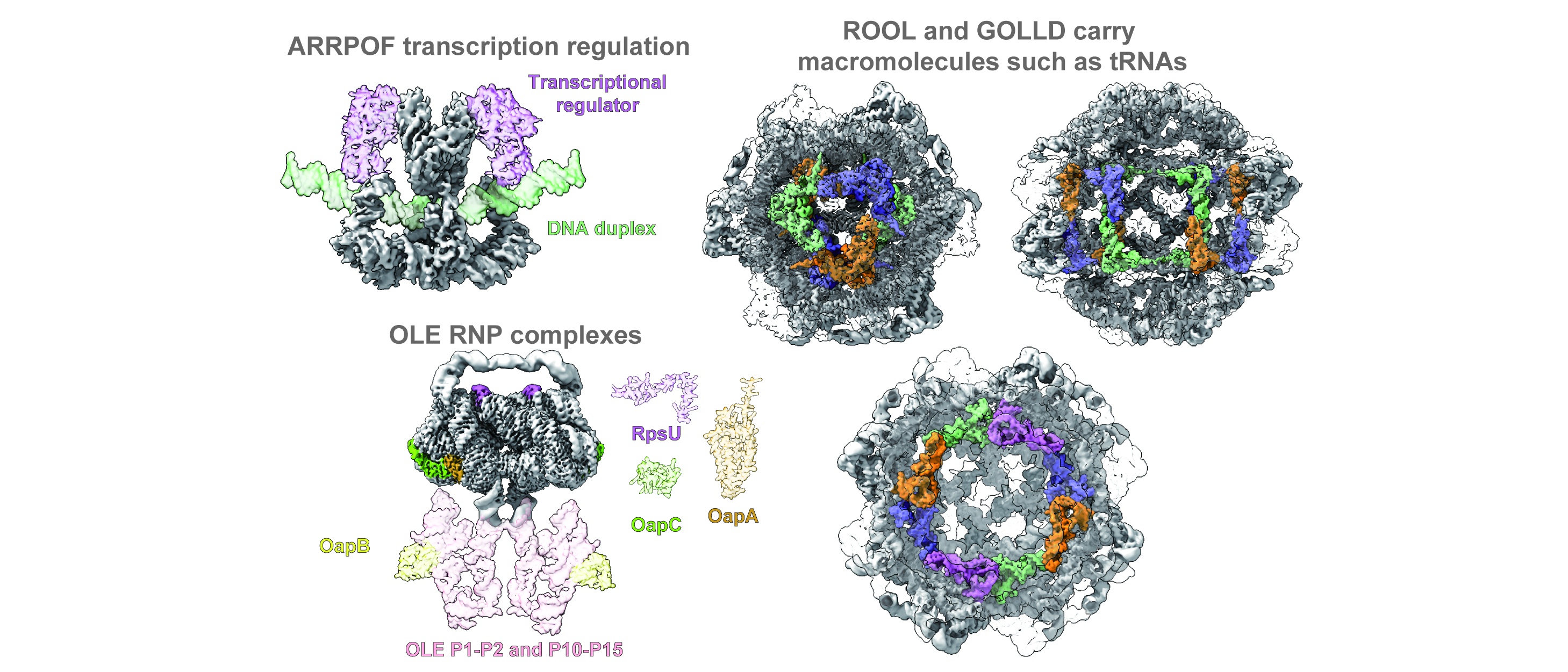
53. Wang, L.#, Xie, J.#, Gong, T.#, Wu, H.#, Tu, Y.#, Peng, X.#, Shang, S.#, Jia, X.#, Ma, H.#, Zou, J.#, Xu, S.#, Zheng, X.#, Zhang, D.#, Liu, Y.#, et al, Sun, S.*, Zhou, X.*, Su, Z.*, Cryo-EM reveals mechanisms of natural RNA multivalency. Science, 2025, adv3451.
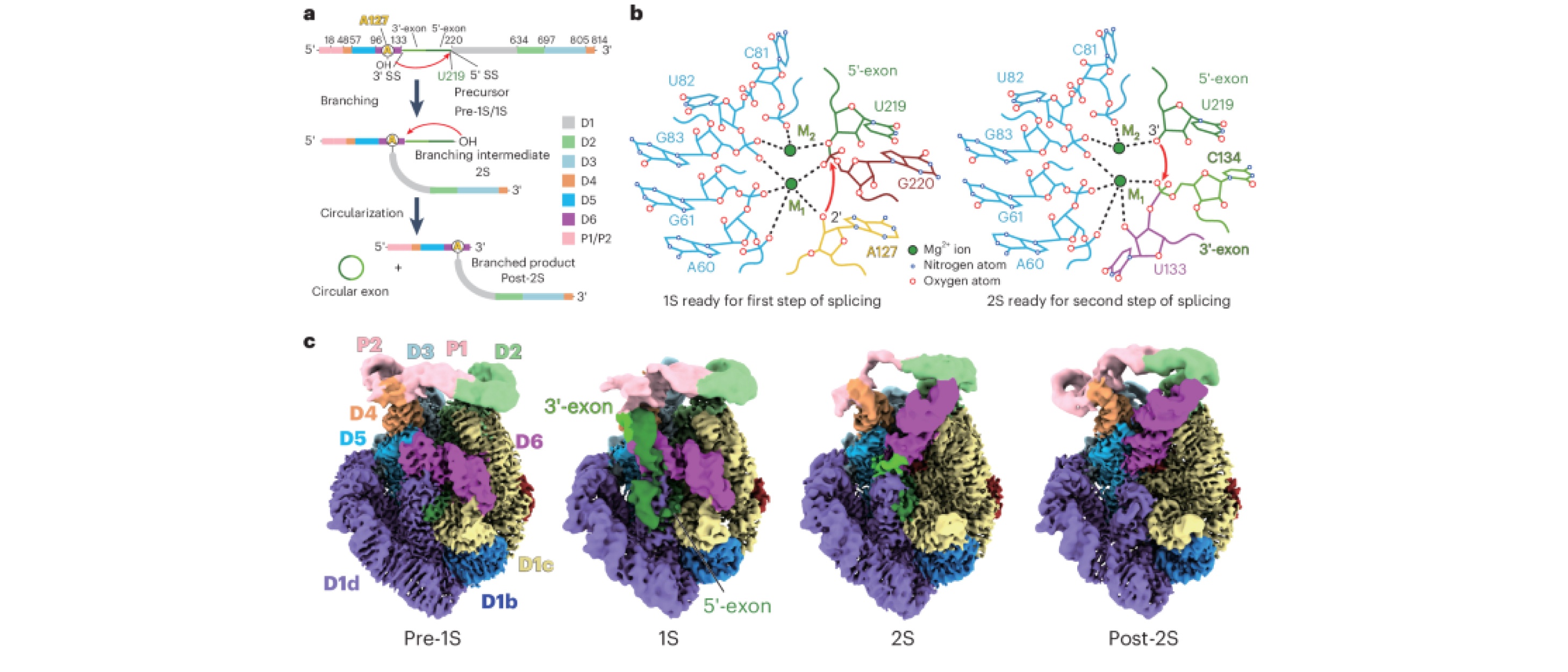
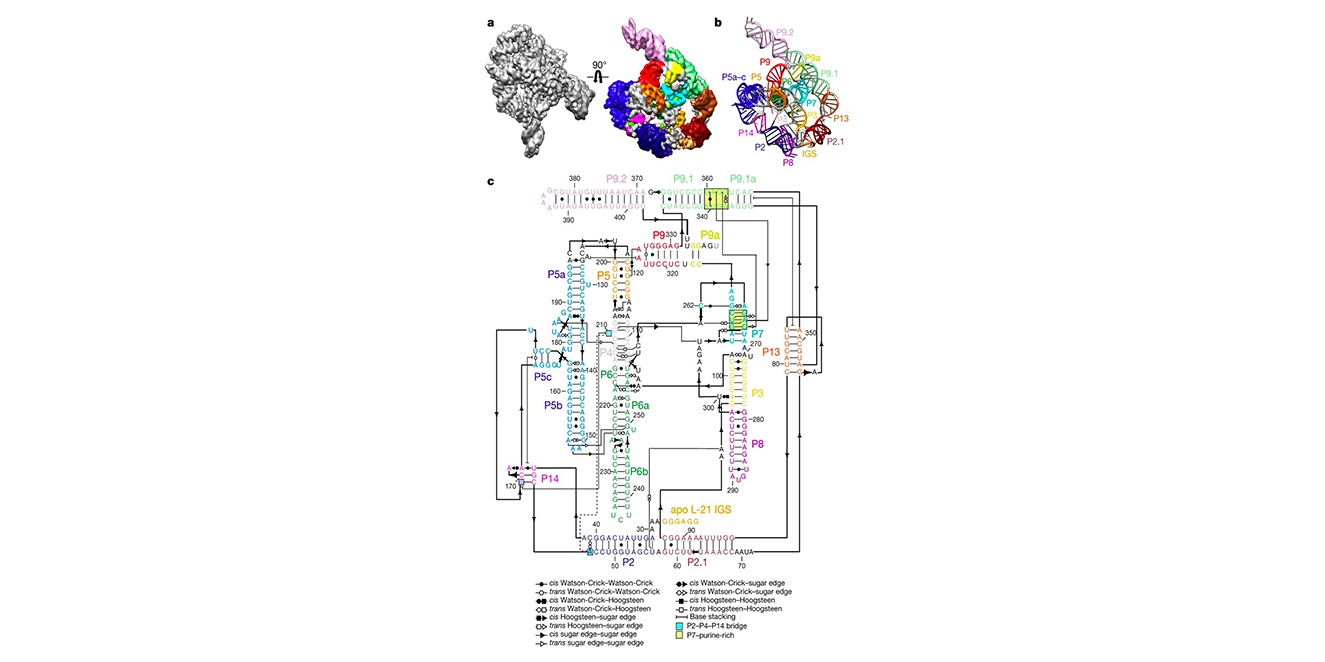
52. Wang, L.#, Xie, J.#, Zhang, C.#, Zou, J., Huang, Z., Shang, S., Chen, X., Yang, Y., Liu, J., Dong, H., Huang, D.*, Su, Z.* Structural basis of circularly permuted group II intron self-splicing. Nat. Struct. Mol. Biol., 2025, 1-10.
30. Jia, X.#, Pan, Z.#, Yuan, Y.#, Luo, B., Luo, Y., Mukherjee, S., Jia, G., Liu, L., Ling, X., Yang, X., Miao, Z., Wei, X., Bjnicki, J. M., Zhao, K.* & Su, Z.* Structural basis of sRNA RsmZ regulation of Pseudomonas aeruginosa virulence. Cell Res., 2023, 33:328-330

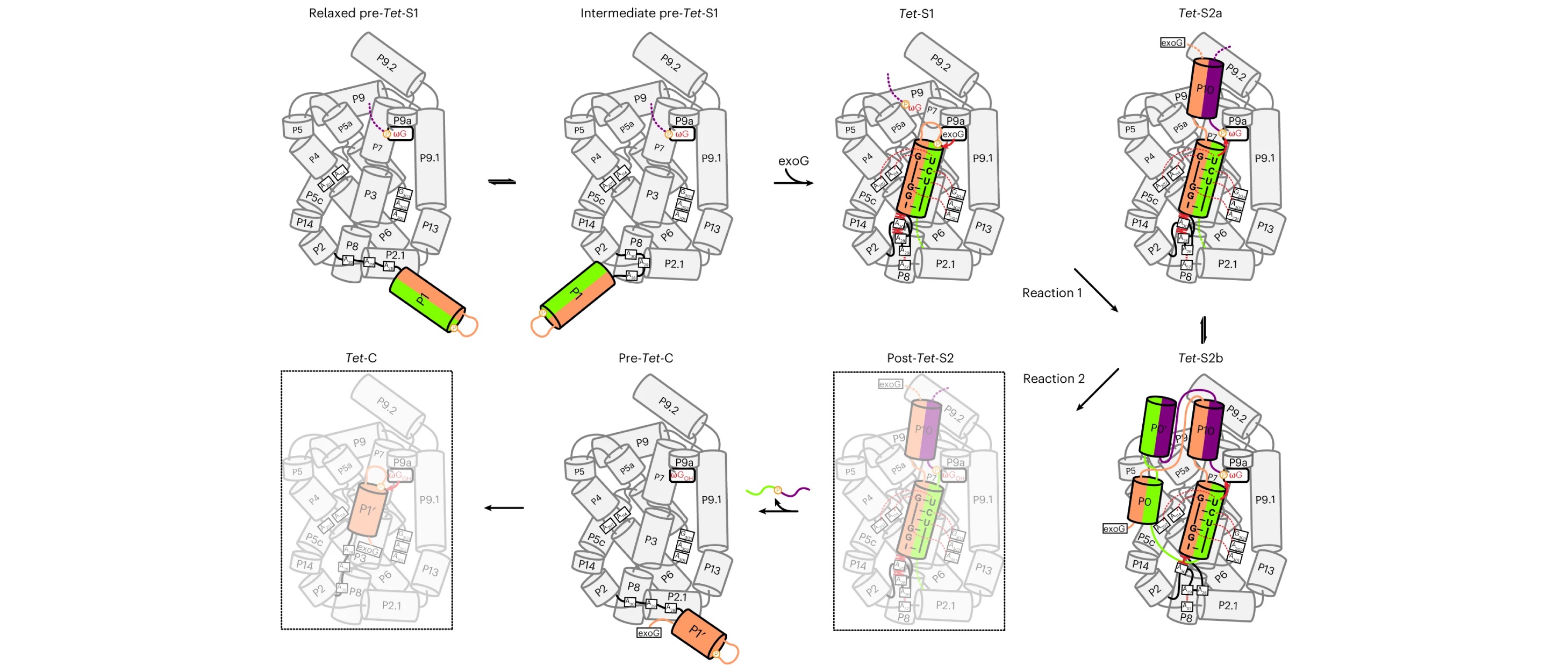
29. Luo, B.# , Zhang, C.# , Ling, X., Mukherjee, S., Jia, G., Xie, J., Jia, X., Liu, L., Luo, Y., Jiang, L., Dong, H., Wei, X., Bujnicki, J.M., Su, Z.*, Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing. Nat. Catal., 2023, 6:298-309.
16. Lyu, J., Yang, M., Zhang, C., Luo, Y., Qin, T., Su, Z., Huang, Z.*, DNA nanostructures directed by RNA clamps. Nanoscale., 2021, 13(47):19870-19874.
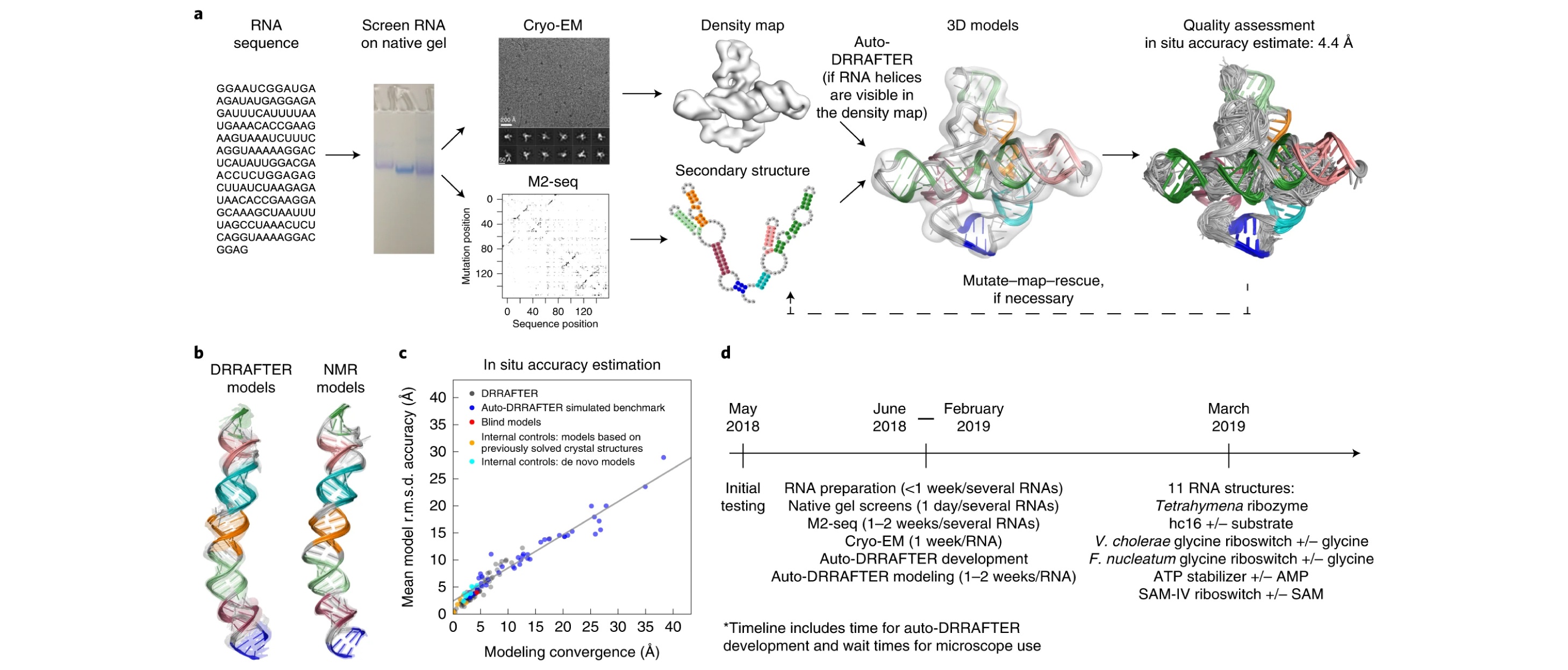
11. Kappel, K.#, Zhang, K.#, Kappel, K.#, Zhang, K.#, Su, Z.#, Watkins, A.M., Kladwang, W., Li, S., Pintilie, G.D., Topkar, V.V., Rangan, R., Zheludev, I.N., Yesselman, J.D., Chiu, W.*, & Das, R.* Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures. Nat.Methods, 2020, 17, 699-707.
9. Zhang, K.#, Li, S.#, Kappel, K.#, Pintilie, G.D., Su, Z., Mou, T.-C., Schimid, M.F., Das, R.*, & Chiu, W.* Cryo-EM structure of 40-kDa SAM-IV riboswitch RNA at 3.7 Å resolution. Nat. Commun. , 2019, 10(1), 1-6.
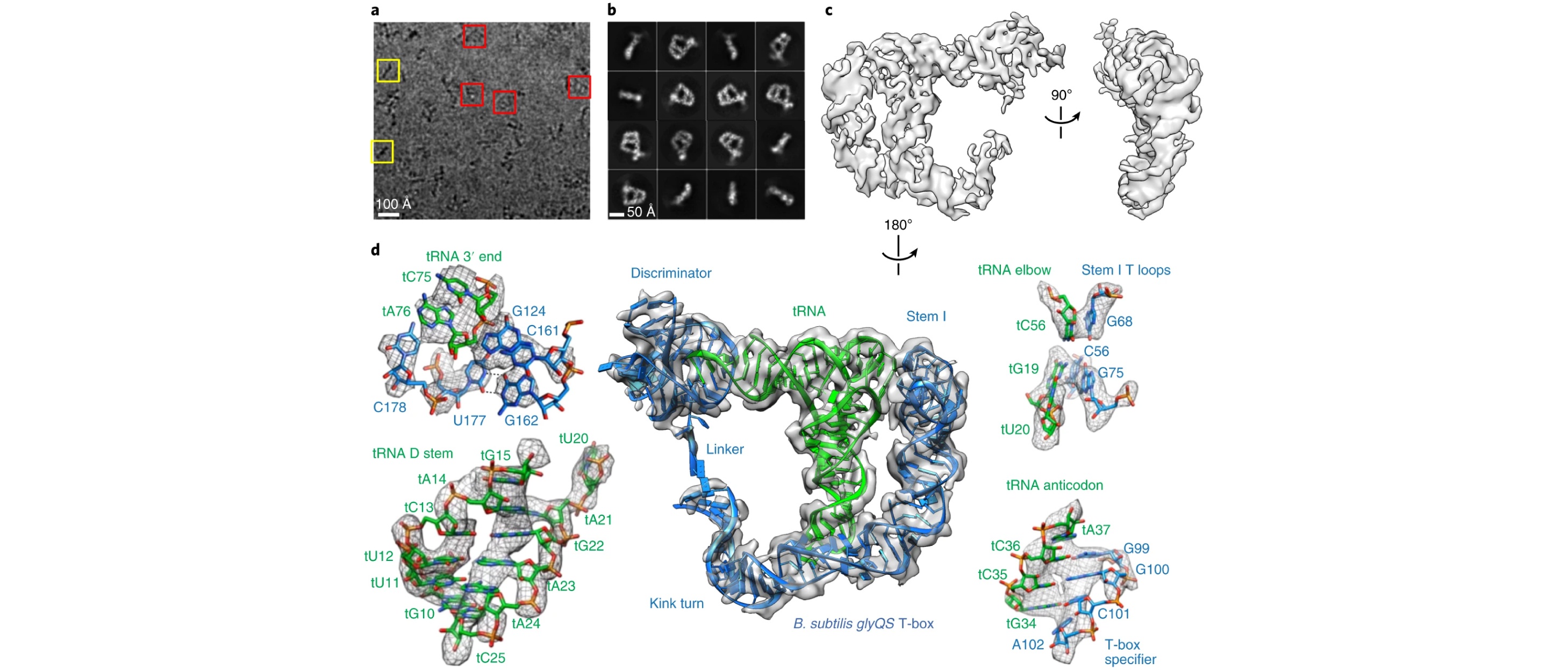
8. Li, S. # , Su, Z. # , et al., Chiu, W.* and Zhang, J.*, Structural basis of amino acid surveillance by higher order tRNA-mRNA interactions. Nat. Struct. Mol. Biol., 2019, 26(12):1094-1105.
6. Su, Z.#, Wu, C.#, Shi, L., Luthra, P., Pintilie, G.D., Britney, J., Porter, J.R., Ge, P., Chen, M., Liu, G., Frederick, T.E., Binning, J.M., Bowman, G.R., Zhou, Z.H., Basler, C.F., Gross, M.L., Leung, D.W., Chiu, W.*, Amarasinghe, G.K.* Electron cryo-microscopy structure of Ebola nucleoprotein reveals a mechanism for nucleocapsid-like assembly. Cell, 2018, 172(5), 966-978.